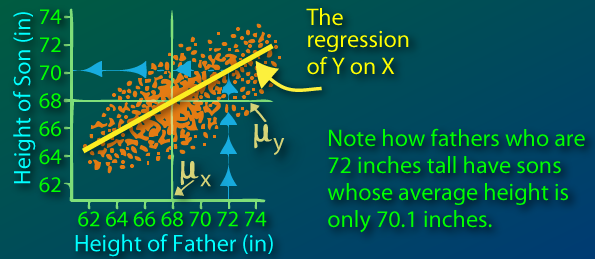
Sir Francis Galton first observed this phenomenon when comparing parents' heights to children's heights, and noticed that while there was a strong correlation between the two, there was also this phenomenon of extreme parent heights leading to less extreme heights in offsprings. As heights from one generation to another became less and less extreme and trended towards some constant, later identified as the mean, the expression "regression toward the mean" appeared.
(from http://www.animatedsoftware.com/statglos/sgregmea.htm)
Now an obvious paradox comes out from this: if all heights become less and less extreme and converge wouldn't we all be of the same height today? How is diversity maintained?
As mentioned I will look at this from a genetics perspective under some simple assumptions.
First model
To kick things off, I will consider a certain trait such as height, and assume that each child will have the average height of his two parents. For simulation purposes, I will consider a population of 100 males, 100 females, and each couple having exactly two children, one boy one girl in order to maintain population size and composition. The physical trait of the original population is generated via a discrete 0-100 uniform distribution.
Let's see how this evolves over 20 generations:
The population average remains constant which makes sense: if the children's height is the average of the parents' height, the average height of the new generation is going to be the same as for the previous generation. We clearly see the regression toward the mean in effect, the speed of convergence is rather fascinating. While we have a min and max of 0 and 100 in the first generation, the entire population has a value between 49 and 55 in the 10th generation.
Our assumption that the physical trait is the exact average of both parents is a stretch and few traits are passed on this way. Let us look at a more biologically-accurate model.
Second model
How is genetic information actually passed on from generation to generation? I will not go into the full details, but we know that we actually inherit two copies of each gene, one from mom one from dad. Assuming their is a gene that directly controls height, dad will send one version of this gene (called allele), and so will mom. Since they themselves have two copies of each, there is a 50/50 chance of which one my dad will give me, same for mom.
(from http://cikgurozaini.blogspot.com/2010/06/genetic-1.html)
So let's take another model which replicates this. Each parent has two alleles (again we draw from a discrete 0-100 uniform distribution) and randomly passes one to the offspring.
We also assume that the value we observe for the individual (phenotype) is the average of these two values.
So if dad has 10 and 50, we will see him as a 30, and if mom is 30 and 90, we will see her as a 60.
What about the offspring? Well, here are the possible outcomes:
- 25% chance the child will be 10 and 30 (phenotype 20)
- 25% chance of being 10 and 90 (phenotype 50)
- 25% chance of being 50 and 30 (phenotype 40)
- 25% chance of being 50 and 90 (phenotype 70)
Now this is where we see the regression to the mean effect: the expected value of the child is 45 (0.25 * 20 + 0.25 * 50 + 0.25 * 40 + 0.25 * 70), which happens to be the parents' average (0.5 * 30 + 0.5 * 60). But this is only an expectation, and diversity is maintained with the child's value possibly ranging from 20 to 70.
So what happens when we repeat the process over and over?
(For a reason I will explain later, I have increased population to 20,000 instead of 200 from the first model)

A thousand generations down the road not much has changed! Looking at the count of unique phenotypes and alleles confirms this population heterogeneity:

The expectation is for children to have their parents' average height, but random mix-and-matching ensure diversity. Also note that mutations can occur, thus creating newer alleles!
This model, much more accurate both from a genetics standpoint and from what we observe in our everyday life, explains the paradox behind the original regression toward the mean: it's not because the expectation is the mean that you will converge towards it.
So there we have it, regression toward the mean does not imply we will all be clones of each other down the road!
Side note on population increase:
At the start of the second modeling phase, I had mention I had increased the size of my population from 200 to 20,000. This is because alleles can disappear in small populations: If dad has alleles a1 and a2 and two kids, the probability that a1 never gets passed on is 25%. So at each new generation, each parental allele has a 25% probability of disappearing Of course allele a1 could be present for another parent who might pass it one, but the fact remains that all alleles are in constant risk of not being passed on. When simulating over many generations, this is actually what happens, and once a given allele disappears within the population, it can never be generated again (forgetting mutations for an instant).
Looking at a population of only 200 individuals:
After 1000 generations, the entire population has converged to a unique height. The loss of alleles (and thus phenotypes) can be viewed here:
The number of unique alleles is decreasing as alleles can disappear but never re-appear. As for the number of phenotypes it can increase. Indeed, suppose mom and dad are both 10-30 (same phenotype of 20), their offspring can be 10-10 (phenotype 10) or 10-30 (phenotype 20) or 30-30 (phenotype 30) so potentially three different phenotypes. The number of phenotypes is nonetheless going to be driven by the number of alleles. The less different alleles there are in the population, the less different combinations you can create.
If we increase the sample size, we decrease the probability of alleles disappearing. It would be quite unlikely for all parents with allele a1 to not pass it down to their children! However, theory around random walk indicates that this is inevitable after sufficient generations, no matter the size of the population (this is what we started to observe in our 20,000 population with the number of alleles starting to decrease around generation 400).